|
| 1 | +# ISIC-2017 Task1 |
| 2 | + |
| 3 | +## Description |
| 4 | + |
| 5 | +This project support **`ISIC-2017 Task1 `**, and the dataset used in this project can be downloaded from [here](https://challenge.isic-archive.com/data/#2017). |
| 6 | + |
| 7 | +### Dataset Overview |
| 8 | + |
| 9 | +The goal of the challenge is to help participants develop image analysis tools to enable the automated diagnosis of melanoma from dermoscopic images. |
| 10 | + |
| 11 | +This challenge provides training data (~2000 images) for participants to engage in all 3 components of lesion image analysis. A separate public validation dataset (~150 images) and blind held-out test dataset (~600 images) will be provided for participants to generate and submit automated results. |
| 12 | + |
| 13 | +### Original Statistic Information |
| 14 | + |
| 15 | +| Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License | |
| 16 | +| ---------------------------------------------------------------- | ----------------- | ------------ | ---------- | ------------ | --------------------- | ---------------------- | ------------ | ---------------------------------------------------------------------- | |
| 17 | +| [ISIC-2017 Task1](https://challenge.isic-archive.com/data/#2017) | full body | segmentation | dermoscopy | 2 | 2000/150/600 | yes/yes/yes | 2017 | [CC-0](https://creativecommons.org/share-your-work/public-domain/cc0/) | |
| 18 | + |
| 19 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 20 | +| :---------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 21 | +| normal | 2000 | 82.86 | 150 | 73.88 | 600 | 70.62 | |
| 22 | +| skin lesion | 2000 | 17.14 | 150 | 26.12 | 600 | 29.38 | |
| 23 | + |
| 24 | +Note: |
| 25 | + |
| 26 | +- `Pct` means percentage of pixels in this category in all pixels. |
| 27 | + |
| 28 | +### Visualization |
| 29 | + |
| 30 | +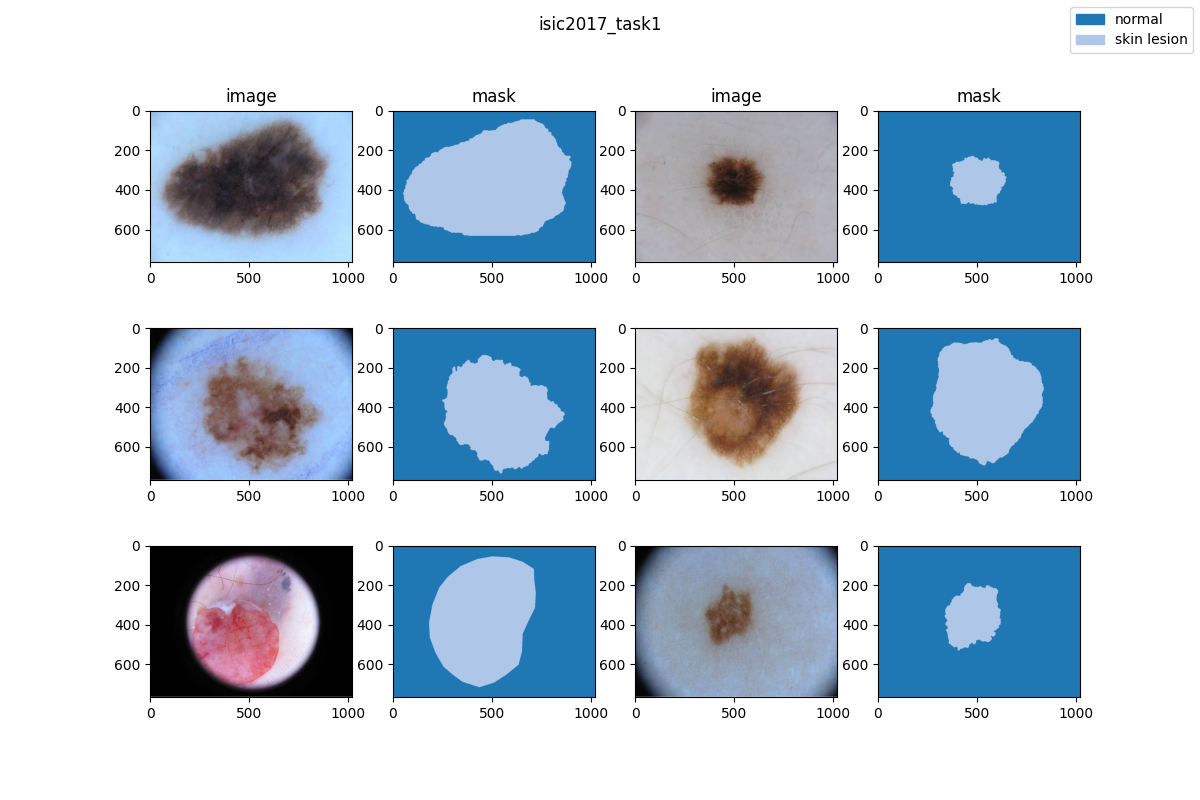 |
| 31 | + |
| 32 | +### Prerequisites |
| 33 | + |
| 34 | +- Python 3.8 |
| 35 | +- PyTorch 1.10.0 |
| 36 | +- pillow(PIL) 9.3.0 |
| 37 | +- scikit-learn(sklearn) 1.2.0 |
| 38 | +- [MIM](https://github.com/open-mmlab/mim) v0.3.4 |
| 39 | +- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 |
| 40 | +- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher |
| 41 | +- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5 |
| 42 | + |
| 43 | +All the commands below rely on the correct configuration of PYTHONPATH, which should point to the project's directory so that Python can locate the module files. In isic2017_task1/ root directory, run the following line to add the current directory to PYTHONPATH: |
| 44 | + |
| 45 | +```shell |
| 46 | +export PYTHONPATH=`pwd`:$PYTHONPATH |
| 47 | +``` |
| 48 | + |
| 49 | +### Dataset preparing |
| 50 | + |
| 51 | +- download dataset from [here](https://challenge.isic-archive.com/data/#2017) and decompression data to path 'data/'. |
| 52 | +- run script `"python tools/prepare_dataset.py"` to split dataset and change folder structure as below. |
| 53 | +- run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt` and `test.txt`. If the label of official validation set and test set can't be obtained, we generate `train.txt` and `val.txt` from the training set randomly. |
| 54 | + |
| 55 | +```none |
| 56 | + mmsegmentation |
| 57 | + ├── mmseg |
| 58 | + ├── projects |
| 59 | + │ ├── medical |
| 60 | + │ │ ├── 2d_image |
| 61 | + │ │ │ ├── dermoscopy |
| 62 | + │ │ │ │ ├── isic2017_task1 |
| 63 | + │ │ │ │ │ ├── configs |
| 64 | + │ │ │ │ │ ├── datasets |
| 65 | + │ │ │ │ │ ├── tools |
| 66 | + │ │ │ │ │ ├── data |
| 67 | + │ │ │ │ │ │ ├── train.txt |
| 68 | + │ │ │ │ │ │ ├── val.txt |
| 69 | + │ │ │ │ │ │ ├── test.txt |
| 70 | + │ │ │ │ │ │ ├── images |
| 71 | + │ │ │ │ │ │ │ ├── train |
| 72 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 73 | + │ │ │ │ | │ │ │ ├── ... |
| 74 | + │ │ │ │ | │ │ │ └── xxx.png |
| 75 | + │ │ │ │ │ │ │ ├── val |
| 76 | + │ │ │ │ | │ │ │ ├── yyy.png |
| 77 | + │ │ │ │ | │ │ │ ├── ... |
| 78 | + │ │ │ │ | │ │ │ └── yyy.png |
| 79 | + │ │ │ │ │ │ │ ├── test |
| 80 | + │ │ │ │ | │ │ │ ├── yyy.png |
| 81 | + │ │ │ │ | │ │ │ ├── ... |
| 82 | + │ │ │ │ | │ │ │ └── yyy.png |
| 83 | + │ │ │ │ │ │ ├── masks |
| 84 | + │ │ │ │ │ │ │ ├── train |
| 85 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 86 | + │ │ │ │ | │ │ │ ├── ... |
| 87 | + │ │ │ │ | │ │ │ └── xxx.png |
| 88 | + │ │ │ │ │ │ │ ├── val |
| 89 | + │ │ │ │ | │ │ │ ├── yyy.png |
| 90 | + │ │ │ │ | │ │ │ ├── ... |
| 91 | + │ │ │ │ | │ │ │ └── yyy.png |
| 92 | + │ │ │ │ │ │ │ ├── test |
| 93 | + │ │ │ │ | │ │ │ ├── yyy.png |
| 94 | + │ │ │ │ | │ │ │ ├── ... |
| 95 | + │ │ │ │ | │ │ │ └── yyy.png |
| 96 | +``` |
| 97 | + |
| 98 | +### Training commands |
| 99 | + |
| 100 | +```shell |
| 101 | +mim train mmseg ./configs/${CONFIG_PATH} |
| 102 | +``` |
| 103 | + |
| 104 | +To train on multiple GPUs, e.g. 8 GPUs, run the following command: |
| 105 | + |
| 106 | +```shell |
| 107 | +mim train mmseg ./configs/${CONFIG_PATH} --launcher pytorch --gpus 8 |
| 108 | +``` |
| 109 | + |
| 110 | +### Testing commands |
| 111 | + |
| 112 | +```shell |
| 113 | +mim test mmseg ./configs/${CONFIG_PATH} --checkpoint ${CHECKPOINT_PATH} |
| 114 | +``` |
| 115 | + |
| 116 | +<!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models) |
| 117 | +
|
| 118 | +You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. --> |
| 119 | + |
| 120 | +## Results |
| 121 | + |
| 122 | +### ISIC-2017 Task1 |
| 123 | + |
| 124 | +| Method | Backbone | Crop Size | lr | mIoU | mDice | config | |
| 125 | +| :-------------: | :------: | :-------: | :----: | :--: | :---: | :---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------: | |
| 126 | +| fcn_unet_s5-d16 | unet | 512x512 | 0.01 | - | - | [config](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/projects/medical/2d_image/dermoscopy/isic2017_task1/configs/fcn-unet-s5-d16_unet_1xb16-0.01-20k_isic2017-task1-512x512.py) | |
| 127 | +| fcn_unet_s5-d16 | unet | 512x512 | 0.001 | - | - | [config](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/projects/medical/2d_image/dermoscopy/isic2017_task1/configs/fcn-unet-s5-d16_unet_1xb16-0.001-20k_isic2017-task1-512x512.py) | |
| 128 | +| fcn_unet_s5-d16 | unet | 512x512 | 0.0001 | - | - | [config](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/projects/medical/2d_image/dermoscopy/isic2017_task1/configs/fcn-unet-s5-d16_unet_1xb16-0.0001-20k_isic2017-task1-512x512.py) | |
| 129 | + |
| 130 | +## Checklist |
| 131 | + |
| 132 | +- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. |
| 133 | + |
| 134 | + - [x] Finish the code |
| 135 | + |
| 136 | + - [x] Basic docstrings & proper citation |
| 137 | + |
| 138 | + - [ ] Test-time correctness |
| 139 | + |
| 140 | + - [x] A full README |
| 141 | + |
| 142 | +- [ ] Milestone 2: Indicates a successful model implementation. |
| 143 | + |
| 144 | + - [ ] Training-time correctness |
| 145 | + |
| 146 | +- [ ] Milestone 3: Good to be a part of our core package! |
| 147 | + |
| 148 | + - [ ] Type hints and docstrings |
| 149 | + |
| 150 | + - [ ] Unit tests |
| 151 | + |
| 152 | + - [ ] Code polishing |
| 153 | + |
| 154 | + - [ ] Metafile.yml |
| 155 | + |
| 156 | +- [ ] Move your modules into the core package following the codebase's file hierarchy structure. |
| 157 | + |
| 158 | +- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
0 commit comments