|
| 1 | +# Chest Radiograph Anatomical Structure Segmentation (CRASS) |
| 2 | + |
| 3 | +## Description |
| 4 | + |
| 5 | +This project supports **`Chest Radiograph Anatomical Structure Segmentation (CRASS) `**, which can be downloaded from [here](https://crass.grand-challenge.org/). |
| 6 | + |
| 7 | +### Dataset Overview |
| 8 | + |
| 9 | +A set of consecutively obtained posterior-anterior chest radiograph were selected from a database containing images acquired at two sites in sub Saharan Africa with a high tuberculosis incidence. All subjects were 15 years or older. Images from digital chest radiography units were used (Delft Imaging Systems, The Netherlands) of varying resolutions, with a typical resolution of 1800--2000 pixels, the pixel size was 250 lm isotropic. From the total set of images, 225 were considered to be normal by an expert radiologist, while 333 of the images contained abnormalities. Of the abnormal images, 220 contained abnormalities in the upper area of the lung where the clavicle is located. The data was divided into a training and a test set. The training set consisted of 299 images, the test set of 249 images. |
| 10 | +The current data is still incomplete and to be added later. |
| 11 | + |
| 12 | +### Information Statistics |
| 13 | + |
| 14 | +| Dataset Name | Anatomical Region | Task Type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License | |
| 15 | +| ------------------------------------------- | ----------------- | ------------ | -------- | ------------ | --------------------- | ---------------------- | ------------ | ------------------------------------------------------------- | |
| 16 | +| [crass](https://crass.grand-challenge.org/) | pulmonary | segmentation | x_ray | 2 | 299/-/234 | yes/-/no | 2021 | [CC0 1.0](https://creativecommons.org/publicdomain/zero/1.0/) | |
| 17 | + |
| 18 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 19 | +| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 20 | +| background | 299 | 98.38 | - | - | - | - | |
| 21 | +| clavicles | 299 | 1.62 | - | - | - | - | |
| 22 | + |
| 23 | +Note: |
| 24 | + |
| 25 | +- `Pct` means percentage of pixels in this category in all pixels. |
| 26 | + |
| 27 | +### Visualization |
| 28 | + |
| 29 | +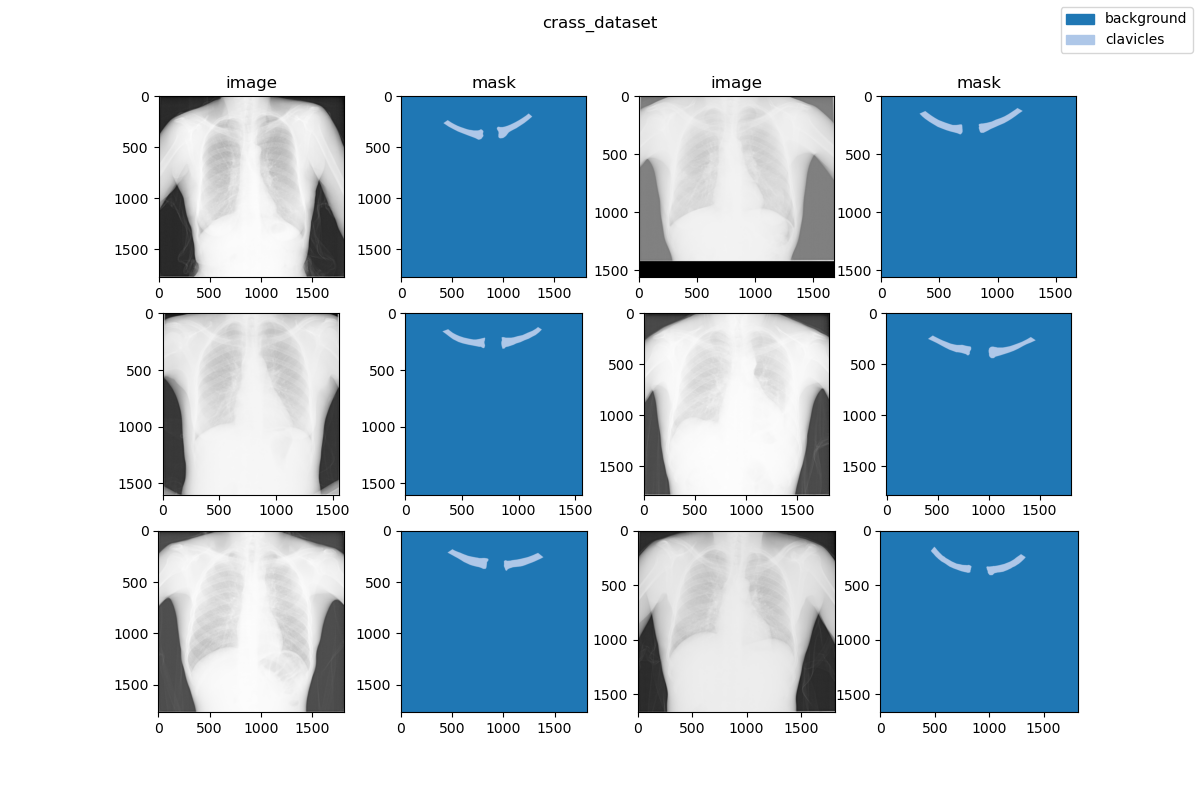 |
| 30 | + |
| 31 | +### Dataset Citation |
| 32 | + |
| 33 | +``` |
| 34 | +@article{HOGEWEG20121490, |
| 35 | + title={Clavicle segmentation in chest radiographs}, |
| 36 | + journal={Medical Image Analysis}, |
| 37 | + volume={16}, |
| 38 | + number={8}, |
| 39 | + pages={1490-1502}, |
| 40 | + year={2012} |
| 41 | +} |
| 42 | +``` |
| 43 | + |
| 44 | +### Prerequisites |
| 45 | + |
| 46 | +- Python v3.8 |
| 47 | +- PyTorch v1.10.0 |
| 48 | +- pillow(PIL) v9.3.0 |
| 49 | +- scikit-learn(sklearn) v1.2.0 |
| 50 | +- [MIM](https://github.com/open-mmlab/mim) v0.3.4 |
| 51 | +- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 |
| 52 | +- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher |
| 53 | +- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5 |
| 54 | + |
| 55 | +All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `crass/` root directory, run the following line to add the current directory to `PYTHONPATH`: |
| 56 | + |
| 57 | +```shell |
| 58 | +export PYTHONPATH=`pwd`:$PYTHONPATH |
| 59 | +``` |
| 60 | + |
| 61 | +### Dataset Preparing |
| 62 | + |
| 63 | +- download dataset from [here](https://crass.grand-challenge.org/) and decompress data to path `'data/'`. |
| 64 | +- run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below. |
| 65 | +- run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set cannot be obtained, we generate `train.txt` and `val.txt` from the training set randomly. |
| 66 | + |
| 67 | +```none |
| 68 | + mmsegmentation |
| 69 | + ├── mmseg |
| 70 | + ├── projects |
| 71 | + │ ├── medical |
| 72 | + │ │ ├── 2d_image |
| 73 | + │ │ │ ├── x_ray |
| 74 | + │ │ │ │ ├── crass |
| 75 | + │ │ │ │ │ ├── configs |
| 76 | + │ │ │ │ │ ├── datasets |
| 77 | + │ │ │ │ │ ├── tools |
| 78 | + │ │ │ │ │ ├── data |
| 79 | + │ │ │ │ │ │ ├── train.txt |
| 80 | + │ │ │ │ │ │ ├── val.txt |
| 81 | + │ │ │ │ │ │ ├── images |
| 82 | + │ │ │ │ │ │ │ ├── train |
| 83 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 84 | + │ │ │ │ | │ │ │ ├── ... |
| 85 | + │ │ │ │ | │ │ │ └── xxx.png |
| 86 | + │ │ │ │ │ │ ├── masks |
| 87 | + │ │ │ │ │ │ │ ├── train |
| 88 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 89 | + │ │ │ │ | │ │ │ ├── ... |
| 90 | + │ │ │ │ | │ │ │ └── xxx.png |
| 91 | +``` |
| 92 | + |
| 93 | +### Divided Dataset Information |
| 94 | + |
| 95 | +***Note: The table information below is divided by ourselves.*** |
| 96 | + |
| 97 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 98 | +| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 99 | +| background | 227 | 98.38 | 57 | 98.39 | - | - | |
| 100 | +| clavicles | 227 | 1.62 | 57 | 1.61 | - | - | |
| 101 | + |
| 102 | +### Training commands |
| 103 | + |
| 104 | +To train models on a single server with one GPU. (default) |
| 105 | + |
| 106 | +```shell |
| 107 | +mim train mmseg ./configs/${CONFIG_FILE} |
| 108 | +``` |
| 109 | + |
| 110 | +### Testing commands |
| 111 | + |
| 112 | +To test models on a single server with one GPU. (default) |
| 113 | + |
| 114 | +```shell |
| 115 | +mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH} |
| 116 | +``` |
| 117 | + |
| 118 | +<!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models) |
| 119 | +
|
| 120 | +You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. --> |
| 121 | + |
| 122 | +## Checklist |
| 123 | + |
| 124 | +- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. |
| 125 | + |
| 126 | + - [x] Finish the code |
| 127 | + - [x] Basic docstrings & proper citation |
| 128 | + - [ ] Test-time correctness |
| 129 | + - [x] A full README |
| 130 | + |
| 131 | +- [x] Milestone 2: Indicates a successful model implementation. |
| 132 | + |
| 133 | + - [x] Training-time correctness |
| 134 | + |
| 135 | +- [ ] Milestone 3: Good to be a part of our core package! |
| 136 | + |
| 137 | + - [ ] Type hints and docstrings |
| 138 | + - [ ] Unit tests |
| 139 | + - [ ] Code polishing |
| 140 | + - [ ] Metafile.yml |
| 141 | + |
| 142 | +- [ ] Move your modules into the core package following the codebase's file hierarchy structure. |
| 143 | + |
| 144 | +- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
0 commit comments