|
| 1 | +# Chest X-ray Images with Pneumothorax Masks |
| 2 | + |
| 3 | +## Description |
| 4 | + |
| 5 | +This project support **`Chest X-ray Images with Pneumothorax Masks `**, and the dataset used in this project can be downloaded from [here](https://www.kaggle.com/datasets/vbookshelf/pneumothorax-chest-xray-images-and-masks). |
| 6 | + |
| 7 | +### Dataset Overview |
| 8 | + |
| 9 | +A pneumothorax (noo-moe-THOR-aks) is a collapsed lung. A pneumothorax occurs when air leaks into the space between your lung and chest wall. This air pushes on the outside of your lung and makes it collapse. Pneumothorax can be a complete lung collapse or a collapse of only a portion of the lung. |
| 10 | + |
| 11 | +A pneumothorax can be caused by a blunt or penetrating chest injury, certain medical procedures, or damage from underlying lung disease. Or it may occur for no obvious reason. Symptoms usually include sudden chest pain and shortness of breath. On some occasions, a collapsed lung can be a life-threatening event. |
| 12 | + |
| 13 | +Treatment for a pneumothorax usually involves inserting a needle or chest tube between the ribs to remove the excess air. However, a small pneumothorax may heal on its own. |
| 14 | + |
| 15 | +### Statistic Information |
| 16 | + |
| 17 | +| Dataset Name | Anatomical Region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release date | License | |
| 18 | +| --------------------------------------------------------------------------------------------------------------------------------- | ----------------- | ------------ | -------- | ------------ | --------------------- | ---------------------- | ------------ | --------------------------------------------------------------- | |
| 19 | +| [Chest-x-ray-images-with-pneumothorax-masks](https://www.kaggle.com/datasets/vbookshelf/pneumothorax-chest-xray-images-and-masks) | throax | segmentation | x_ray | 2 | 10675/-/1372 | yes/-/yes | 2020 | [CC-BY-NC 4.0](https://creativecommons.org/licenses/by-sa/4.0/) | |
| 20 | + |
| 21 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 22 | +| :----------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 23 | +| background | 10675 | 99.7 | - | - | 1372 | 99.71 | |
| 24 | +| pneumothroax | 2379 | 0.3 | - | - | 290 | 0.29 | |
| 25 | + |
| 26 | +### Visualization |
| 27 | + |
| 28 | +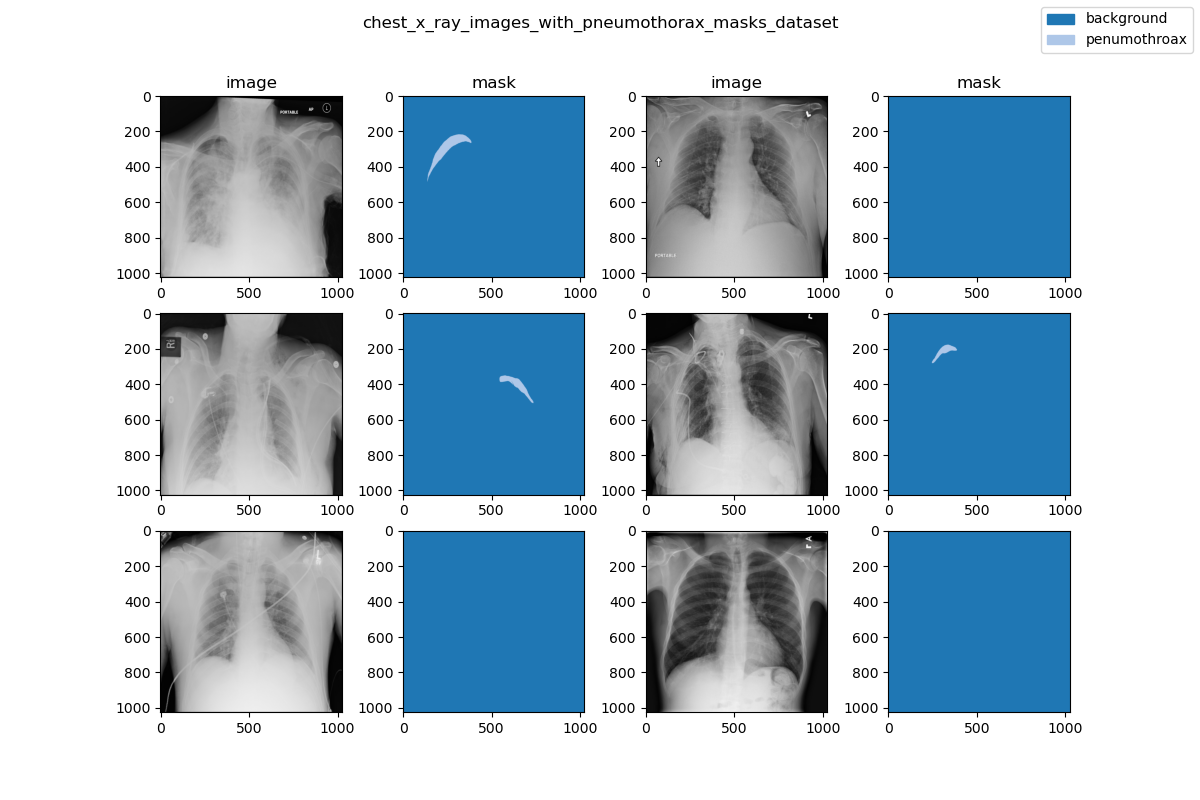 |
| 29 | + |
| 30 | +### Prerequisites |
| 31 | + |
| 32 | +- Python 3.8 |
| 33 | +- PyTorch 1.10.0 |
| 34 | +- pillow(PIL) 9.3.0 |
| 35 | +- scikit-learn(sklearn) 1.2.0 |
| 36 | +- [MIM](https://github.com/open-mmlab/mim) v0.3.4 |
| 37 | +- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 |
| 38 | +- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher |
| 39 | +- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5 |
| 40 | + |
| 41 | +All the commands below rely on the correct configuration of PYTHONPATH, which should point to the project's directory so that Python can locate the module files. In chest_x_ray_images_with_pneumothorax_masks/ root directory, run the following line to add the current directory to PYTHONPATH: |
| 42 | + |
| 43 | +```shell |
| 44 | +export PYTHONPATH=`pwd`:$PYTHONPATH |
| 45 | +``` |
| 46 | + |
| 47 | +### Dataset preparing |
| 48 | + |
| 49 | +- download dataset from [here](https://www.kaggle.com/datasets/vbookshelf/pneumothorax-chest-xray-images-and-masks) and decompression data to path 'data/'. |
| 50 | +- run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below. |
| 51 | +- run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set cannot be obtained, we generate `train.txt` and `val.txt` from the training set randomly. |
| 52 | + |
| 53 | +```none |
| 54 | + mmsegmentation |
| 55 | + ├── mmseg |
| 56 | + ├── projects |
| 57 | + │ ├── medical |
| 58 | + │ │ ├── 2d_image |
| 59 | + │ │ │ ├── x_ray |
| 60 | + │ │ │ │ ├── chest_x_ray_images_with_pneumothorax_masks |
| 61 | + │ │ │ │ │ ├── configs |
| 62 | + │ │ │ │ │ ├── datasets |
| 63 | + │ │ │ │ │ ├── tools |
| 64 | + │ │ │ │ │ ├── data |
| 65 | + │ │ │ │ │ │ ├── train.txt |
| 66 | + │ │ │ │ │ │ ├── val.txt |
| 67 | + │ │ │ │ │ │ ├── images |
| 68 | + │ │ │ │ │ │ │ ├── train |
| 69 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 70 | + │ │ │ │ | │ │ │ ├── ... |
| 71 | + │ │ │ │ | │ │ │ └── xxx.png |
| 72 | + │ │ │ │ │ │ ├── masks |
| 73 | + │ │ │ │ │ │ │ ├── train |
| 74 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 75 | + │ │ │ │ | │ │ │ ├── ... |
| 76 | + │ │ │ │ | │ │ │ └── xxx.png |
| 77 | +``` |
| 78 | + |
| 79 | +### Training commands |
| 80 | + |
| 81 | +```shell |
| 82 | +mim train mmseg ./configs/${CONFIG_PATH} |
| 83 | +``` |
| 84 | + |
| 85 | +To train on multiple GPUs, e.g. 8 GPUs, run the following command: |
| 86 | + |
| 87 | +```shell |
| 88 | +mim train mmseg ./configs/${CONFIG_PATH} --launcher pytorch --gpus 8 |
| 89 | +``` |
| 90 | + |
| 91 | +### Testing commands |
| 92 | + |
| 93 | +```shell |
| 94 | +mim test mmseg ./configs/${CONFIG_PATH} --checkpoint ${CHECKPOINT_PATH} |
| 95 | +``` |
| 96 | + |
| 97 | +## Checklist |
| 98 | + |
| 99 | +- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. |
| 100 | + |
| 101 | + - [x] Finish the code |
| 102 | + - [x] Basic docstrings & proper citation |
| 103 | + - [x] Test-time correctness |
| 104 | + - [x] A full README |
| 105 | + |
| 106 | +- [x] Milestone 2: Indicates a successful model implementation. |
| 107 | + |
| 108 | + - [x] Training-time correctness |
| 109 | + |
| 110 | +- [ ] Milestone 3: Good to be a part of our core package! |
| 111 | + |
| 112 | + - [ ] Type hints and docstrings |
| 113 | + - [ ] Unit tests |
| 114 | + - [ ] Code polishing |
| 115 | + - [ ] Metafile.yml |
| 116 | + |
| 117 | +- [ ] Move your modules into the core package following the codebase's file hierarchy structure. |
| 118 | + |
| 119 | +- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
0 commit comments