|
| 1 | +# Covid-19 CT Chest X-ray Dataset |
| 2 | + |
| 3 | +## Description |
| 4 | + |
| 5 | +This project supports **`Covid-19 CT Chest X-ray Dataset`**, which can be downloaded from [here](https://github.com/ieee8023/covid-chestxray-dataset). |
| 6 | + |
| 7 | +### Dataset Overview |
| 8 | + |
| 9 | +In the context of a COVID-19 pandemic, we want to improve prognostic predictions to triage and manage patient care. Data is the first step to developing any diagnostic/prognostic tool. While there exist large public datasets of more typical chest X-rays from the NIH \[Wang 2017\], Spain \[Bustos 2019\], Stanford \[Irvin 2019\], MIT \[Johnson 2019\] and Indiana University \[Demner-Fushman 2016\], there is no collection of COVID-19 chest X-rays or CT scans designed to be used for computational analysis. |
| 10 | + |
| 11 | +The 2019 novel coronavirus (COVID-19) presents several unique features [Fang, 2020](https://pubs.rsna.org/doi/10.1148/radiol.2020200432) and [Ai 2020](https://pubs.rsna.org/doi/10.1148/radiol.2020200642). While the diagnosis is confirmed using polymerase chain reaction (PCR), infected patients with pneumonia may present on chest X-ray and computed tomography (CT) images with a pattern that is only moderately characteristic for the human eye [Ng, 2020](https://pubs.rsna.org/doi/10.1148/ryct.2020200034). In late January, a Chinese team published a paper detailing the clinical and paraclinical features of COVID-19. They reported that patients present abnormalities in chest CT images with most having bilateral involvement [Huang 2020](<https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30183-5/fulltext>). Bilateral multiple lobular and subsegmental areas of consolidation constitute the typical findings in chest CT images of intensive care unit (ICU) patients on admission [Huang 2020](<https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30183-5/fulltext>). In comparison, non-ICU patients show bilateral ground-glass opacity and subsegmental areas of consolidation in their chest CT images [Huang 2020](<https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30183-5/fulltext>). In these patients, later chest CT images display bilateral ground-glass opacity with resolved consolidation [Huang 2020](<https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30183-5/fulltext>). |
| 12 | + |
| 13 | +### Statistic Information |
| 14 | + |
| 15 | +| Dataset Name | Anatomical Region | Task Type | Modality | Nnum. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release date | License | |
| 16 | +| ---------------------------------------------------------------------- | ----------------- | ------------ | -------- | ------------- | --------------------- | ---------------------- | ------------ | --------------------------------------------------------------------- | |
| 17 | +| [Covid-19-ct-cxr](https://github.com/ieee8023/covid-chestxray-dataset) | thorax | segmentation | x_ray | 2 | 205/-/714 | yes/-/no | 2021 | [CC-BY-NC-SA 4.0](https://creativecommons.org/licenses/by-nc-sa/4.0/) | |
| 18 | + |
| 19 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 20 | +| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 21 | +| background | 205 | 72.84 | - | - | - | - | |
| 22 | +| lung | 205 | 27.16 | - | - | - | - | |
| 23 | + |
| 24 | +Note: |
| 25 | + |
| 26 | +- `Pct` means percentage of pixels in this category in all pixels. |
| 27 | + |
| 28 | +### Visualization |
| 29 | + |
| 30 | +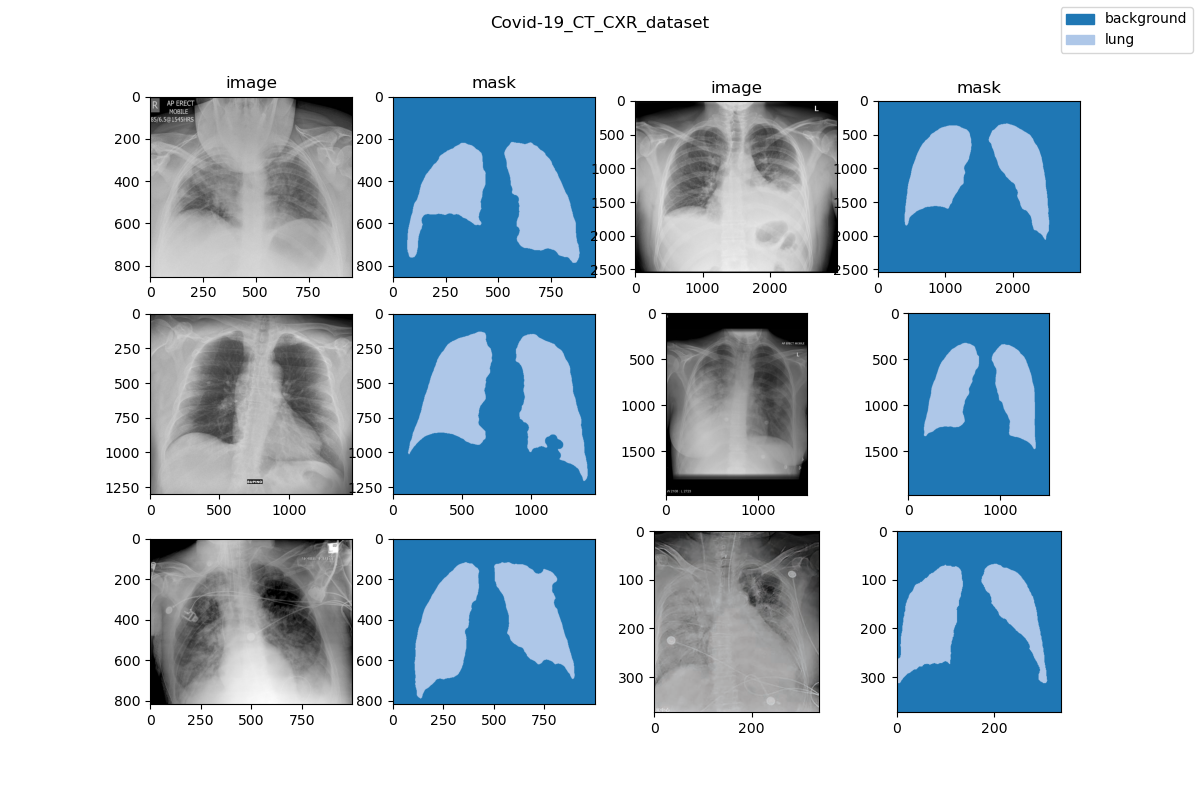 |
| 31 | + |
| 32 | +### Dataset Citation |
| 33 | + |
| 34 | +``` |
| 35 | +@article{cohen2020covidProspective, |
| 36 | + title={{COVID-19} Image Data Collection: Prospective Predictions Are the Future}, |
| 37 | + author={Joseph Paul Cohen and Paul Morrison and Lan Dao and Karsten Roth and Tim Q Duong and Marzyeh Ghassemi}, |
| 38 | + journal={arXiv 2006.11988}, |
| 39 | + year={2020} |
| 40 | +} |
| 41 | +
|
| 42 | +@article{cohen2020covid, |
| 43 | + title={COVID-19 image data collection}, |
| 44 | + author={Joseph Paul Cohen and Paul Morrison and Lan Dao}, |
| 45 | + journal={arXiv 2003.11597}, |
| 46 | + year={2020} |
| 47 | +} |
| 48 | +``` |
| 49 | + |
| 50 | +### Prerequisites |
| 51 | + |
| 52 | +- Python v3.8 |
| 53 | +- PyTorch v1.10.0 |
| 54 | +- pillow(PIL) v9.3.0 9.3.0 |
| 55 | +- scikit-learn(sklearn) v1.2.0 1.2.0 |
| 56 | +- [MIM](https://github.com/open-mmlab/mim) v0.3.4 |
| 57 | +- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 |
| 58 | +- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher |
| 59 | +- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5 |
| 60 | + |
| 61 | +All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `covid_19_ct_cxr/` root directory, run the following line to add the current directory to `PYTHONPATH`: |
| 62 | + |
| 63 | +```shell |
| 64 | +export PYTHONPATH=`pwd`:$PYTHONPATH |
| 65 | +``` |
| 66 | + |
| 67 | +### Dataset Preparing |
| 68 | + |
| 69 | +- download dataset from [here](https://github.com/ieee8023/covid-chestxray-dataset) and decompress data to path `'data/'`. |
| 70 | +- run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below. |
| 71 | +- run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set cannot be obtained, we generate `train.txt` and `val.txt` from the training set randomly. |
| 72 | + |
| 73 | +```shell |
| 74 | +mkdir data && cd data |
| 75 | +git clone [email protected]:ieee8023/covid-chestxray-dataset.git |
| 76 | +cd .. |
| 77 | +python tools/prepare_dataset.py |
| 78 | +python ../../tools/split_seg_dataset.py |
| 79 | +``` |
| 80 | + |
| 81 | +```none |
| 82 | + mmsegmentation |
| 83 | + ├── mmseg |
| 84 | + ├── projects |
| 85 | + │ ├── medical |
| 86 | + │ │ ├── 2d_image |
| 87 | + │ │ │ ├── x_ray |
| 88 | + │ │ │ │ ├── covid_19_ct_cxr |
| 89 | + │ │ │ │ │ ├── configs |
| 90 | + │ │ │ │ │ ├── datasets |
| 91 | + │ │ │ │ │ ├── tools |
| 92 | + │ │ │ │ │ ├── data |
| 93 | + │ │ │ │ │ │ ├── train.txt |
| 94 | + │ │ │ │ │ │ ├── val.txt |
| 95 | + │ │ │ │ │ │ ├── images |
| 96 | + │ │ │ │ │ │ │ ├── train |
| 97 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 98 | + │ │ │ │ | │ │ │ ├── ... |
| 99 | + │ │ │ │ | │ │ │ └── xxx.png |
| 100 | + │ │ │ │ │ │ ├── masks |
| 101 | + │ │ │ │ │ │ │ ├── train |
| 102 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 103 | + │ │ │ │ | │ │ │ ├── ... |
| 104 | + │ │ │ │ | │ │ │ └── xxx.png |
| 105 | +``` |
| 106 | + |
| 107 | +### Divided Dataset Information |
| 108 | + |
| 109 | +***Note: The table information below is divided by ourselves.*** |
| 110 | + |
| 111 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 112 | +| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 113 | +| background | 164 | 72.88 | 41 | 72.69 | - | - | |
| 114 | +| lung | 164 | 27.12 | 41 | 27.31 | - | - | |
| 115 | + |
| 116 | +### Training commands |
| 117 | + |
| 118 | +To train models on a single server with one GPU. (default) |
| 119 | + |
| 120 | +```shell |
| 121 | +mim train mmseg ./configs/${CONFIG_FILE} |
| 122 | +``` |
| 123 | + |
| 124 | +### Testing commands |
| 125 | + |
| 126 | +To test models on a single server with one GPU. (default) |
| 127 | + |
| 128 | +```shell |
| 129 | +mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH} |
| 130 | +``` |
| 131 | + |
| 132 | +<!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models) |
| 133 | +
|
| 134 | +You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. --> |
| 135 | + |
| 136 | +## Checklist |
| 137 | + |
| 138 | +- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. |
| 139 | + |
| 140 | + - [x] Finish the code |
| 141 | + - [x] Basic docstrings & proper citation |
| 142 | + - [x] Test-time correctness |
| 143 | + - [x] A full README |
| 144 | + |
| 145 | +- [x] Milestone 2: Indicates a successful model implementation. |
| 146 | + |
| 147 | + - [x] Training-time correctness |
| 148 | + |
| 149 | +- [ ] Milestone 3: Good to be a part of our core package! |
| 150 | + |
| 151 | + - [ ] Type hints and docstrings |
| 152 | + - [ ] Unit tests |
| 153 | + - [ ] Code polishing |
| 154 | + - [ ] Metafile.yml |
| 155 | + |
| 156 | +- [ ] Move your modules into the core package following the codebase's file hierarchy structure. |
| 157 | + |
| 158 | +- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
0 commit comments