|
| 1 | +# breastCancerCellSegmentation |
| 2 | + |
| 3 | +## Description |
| 4 | + |
| 5 | +This project supports **`breastCancerCellSegmentation`**, which can be downloaded from [here](https://www.heywhale.com/mw/dataset/5e9e9b35ebb37f002c625423). |
| 6 | + |
| 7 | +### Dataset Overview |
| 8 | + |
| 9 | +This dataset, with 58 H&E-stained histopathology images was used for breast cancer cell detection and associated real-world data. |
| 10 | +Conventional histology uses a combination of hematoxylin and eosin stains, commonly referred to as H&E. These images are stained because most cells are inherently transparent with little or no intrinsic pigment. |
| 11 | +Certain special stains selectively bind to specific components and can be used to identify biological structures such as cells. |
| 12 | + |
| 13 | +### Original Statistic Information |
| 14 | + |
| 15 | +| Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License | |
| 16 | +| -------------------------------------------------------------------------------------------- | ----------------- | ------------ | -------------- | ------------ | --------------------- | ---------------------- | ------------ | --------------------------------------------------------------- | |
| 17 | +| [breastCancerCellSegmentation](https://www.heywhale.com/mw/dataset/5e9e9b35ebb37f002c625423) | cell | segmentation | histopathology | 2 | 58/-/- | yes/-/- | 2020 | [CC-BY-NC 4.0](https://creativecommons.org/licenses/by-sa/4.0/) | |
| 18 | + |
| 19 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 20 | +| :--------------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 21 | +| background | 58 | 98.37 | - | - | - | - | |
| 22 | +| breastCancerCell | 58 | 1.63 | - | - | - | - | |
| 23 | + |
| 24 | +Note: |
| 25 | + |
| 26 | +- `Pct` means percentage of pixels in this category in all pixels. |
| 27 | + |
| 28 | +### Visualization |
| 29 | + |
| 30 | +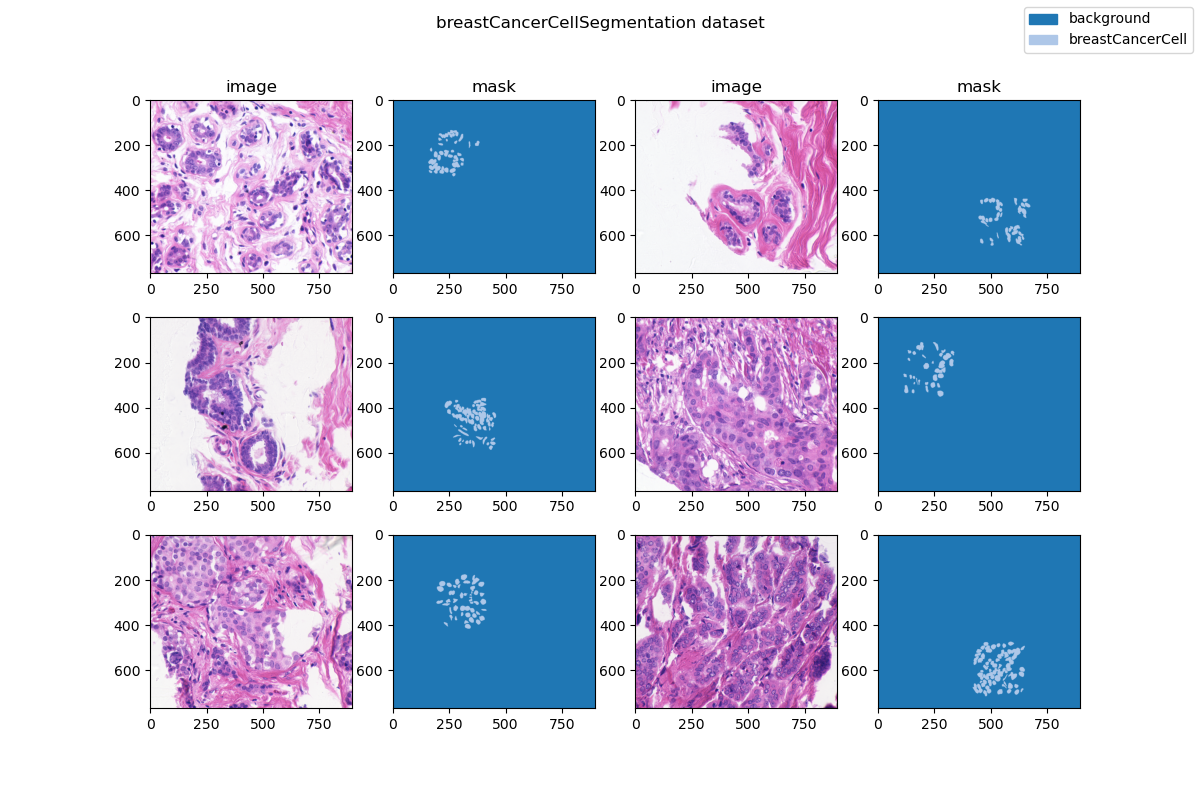 |
| 31 | + |
| 32 | +## Usage |
| 33 | + |
| 34 | +### Prerequisites |
| 35 | + |
| 36 | +- Python v3.8 |
| 37 | +- PyTorch v1.10.0 |
| 38 | +- pillow (PIL) v9.3.0 |
| 39 | +- scikit-learn (sklearn) v1.2.0 |
| 40 | +- [MIM](https://github.com/open-mmlab/mim) v0.3.4 |
| 41 | +- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 |
| 42 | +- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher |
| 43 | +- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0 |
| 44 | + |
| 45 | +All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `breastCancerCellSegmentation/` root directory, run the following line to add the current directory to `PYTHONPATH`: |
| 46 | + |
| 47 | +```shell |
| 48 | +export PYTHONPATH=`pwd`:$PYTHONPATH |
| 49 | +``` |
| 50 | + |
| 51 | +### Dataset Preparing |
| 52 | + |
| 53 | +- Download dataset from [here](https://www.heywhale.com/mw/dataset/5e9e9b35ebb37f002c625423) and save it to the `data/` directory . |
| 54 | +- Decompress data to path `data/`. This will create a new folder named `data/breastCancerCellSegmentation/`, which contains the original image data. |
| 55 | +- run script `python tools/prepare_dataset.py` to format data and change folder structure as below. |
| 56 | + |
| 57 | +```none |
| 58 | + mmsegmentation |
| 59 | + ├── mmseg |
| 60 | + ├── projects |
| 61 | + │ ├── medical |
| 62 | + │ │ ├── 2d_image |
| 63 | + │ │ │ ├── histopathology |
| 64 | + │ │ │ │ ├── breastCancerCellSegmentation |
| 65 | + │ │ │ │ │ ├── configs |
| 66 | + │ │ │ │ │ ├── datasets |
| 67 | + │ │ │ │ │ ├── tools |
| 68 | + │ │ │ │ │ ├── data |
| 69 | + │ │ │ │ │ │ ├── breastCancerCellSegmentation |
| 70 | + | │ │ │ │ │ │ ├── train.txt |
| 71 | + | │ │ │ │ │ │ ├── val.txt |
| 72 | + | │ │ │ │ │ │ ├── images |
| 73 | + | │ │ │ │ │ │ | ├── xxx.tif |
| 74 | + | │ │ │ │ │ │ ├── masks |
| 75 | + | │ │ │ │ │ │ | ├── xxx.TIF |
| 76 | +
|
| 77 | +``` |
| 78 | + |
| 79 | +### Training commands |
| 80 | + |
| 81 | +Train models on a single server with one GPU. |
| 82 | + |
| 83 | +```shell |
| 84 | +mim train mmseg ./configs/${CONFIG_FILE} |
| 85 | +``` |
| 86 | + |
| 87 | +### Testing commands |
| 88 | + |
| 89 | +Test models on a single server with one GPU. |
| 90 | + |
| 91 | +```shell |
| 92 | +mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH} |
| 93 | +``` |
| 94 | + |
| 95 | +## Checklist |
| 96 | + |
| 97 | +- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. |
| 98 | + |
| 99 | + - [x] Finish the code |
| 100 | + |
| 101 | + - [x] Basic docstrings & proper citation |
| 102 | + |
| 103 | + - [x] Test-time correctness |
| 104 | + |
| 105 | + - [x] A full README |
| 106 | + |
| 107 | +- [ ] Milestone 2: Indicates a successful model implementation. |
| 108 | + |
| 109 | + - [ ] Training-time correctness |
| 110 | + |
| 111 | +- [ ] Milestone 3: Good to be a part of our core package! |
| 112 | + |
| 113 | + - [ ] Type hints and docstrings |
| 114 | + |
| 115 | + - [ ] Unit tests |
| 116 | + |
| 117 | + - [ ] Code polishing |
| 118 | + |
| 119 | + - [ ] Metafile.yml |
| 120 | + |
| 121 | +- [ ] Move your modules into the core package following the codebase's file hierarchy structure. |
| 122 | + |
| 123 | +- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
0 commit comments