|
| 1 | +# DR HAGIS: Diabetic Retinopathy, Hypertension, Age-related macular degeneration and Glacuoma ImageS |
| 2 | + |
| 3 | +## Description |
| 4 | + |
| 5 | +This project supports **`DR HAGIS: Diabetic Retinopathy, Hypertension, Age-related macular degeneration and Glacuoma ImageS`**, which can be downloaded from [here](https://paperswithcode.com/dataset/dr-hagis). |
| 6 | + |
| 7 | +### Dataset Overview |
| 8 | + |
| 9 | +The DR HAGIS database has been created to aid the development of vessel extraction algorithms suitable for retinal screening programmes. Researchers are encouraged to test their segmentation algorithms using this database. All thirty-nine fundus images were obtained from a diabetic retinopathy screening programme in the UK. Hence, all images were taken from diabetic patients. |
| 10 | + |
| 11 | +Besides the fundus images, the manual segmentation of the retinal surface vessels is provided by an expert grader. These manually segmented images can be used as the ground truth to compare and assess the automatic vessel extraction algorithms. Masks of the FOV are provided as well to quantify the accuracy of vessel extraction within the FOV only. The images were acquired in different screening centers, therefore reflecting the range of image resolutions, digital cameras and fundus cameras used in the clinic. The fundus images were captured using a Topcon TRC-NW6s, Topcon TRC-NW8 or a Canon CR DGi fundus camera with a horizontal 45 degree field-of-view (FOV). The images are 4752x3168 pixels, 3456x2304 pixels, 3126x2136 pixels, 2896x1944 pixels or 2816x1880 pixels in size. |
| 12 | + |
| 13 | +### Original Statistic Information |
| 14 | + |
| 15 | +| Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License | |
| 16 | +| ------------------------------------------------------- | ----------------- | ------------ | ------------------ | ------------ | --------------------- | ---------------------- | ------------ | ------- | |
| 17 | +| [DR HAGIS](https://paperswithcode.com/dataset/dr-hagis) | head and neck | segmentation | fundus photography | 2 | 40/-/- | yes/-/- | 2017 | - | |
| 18 | + |
| 19 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 20 | +| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 21 | +| background | 40 | 96.38 | - | - | - | - | |
| 22 | +| vessel | 40 | 3.62 | - | - | - | - | |
| 23 | + |
| 24 | +Note: |
| 25 | + |
| 26 | +- `Pct` means percentage of pixels in this category in all pixels. |
| 27 | + |
| 28 | +### Visualization |
| 29 | + |
| 30 | +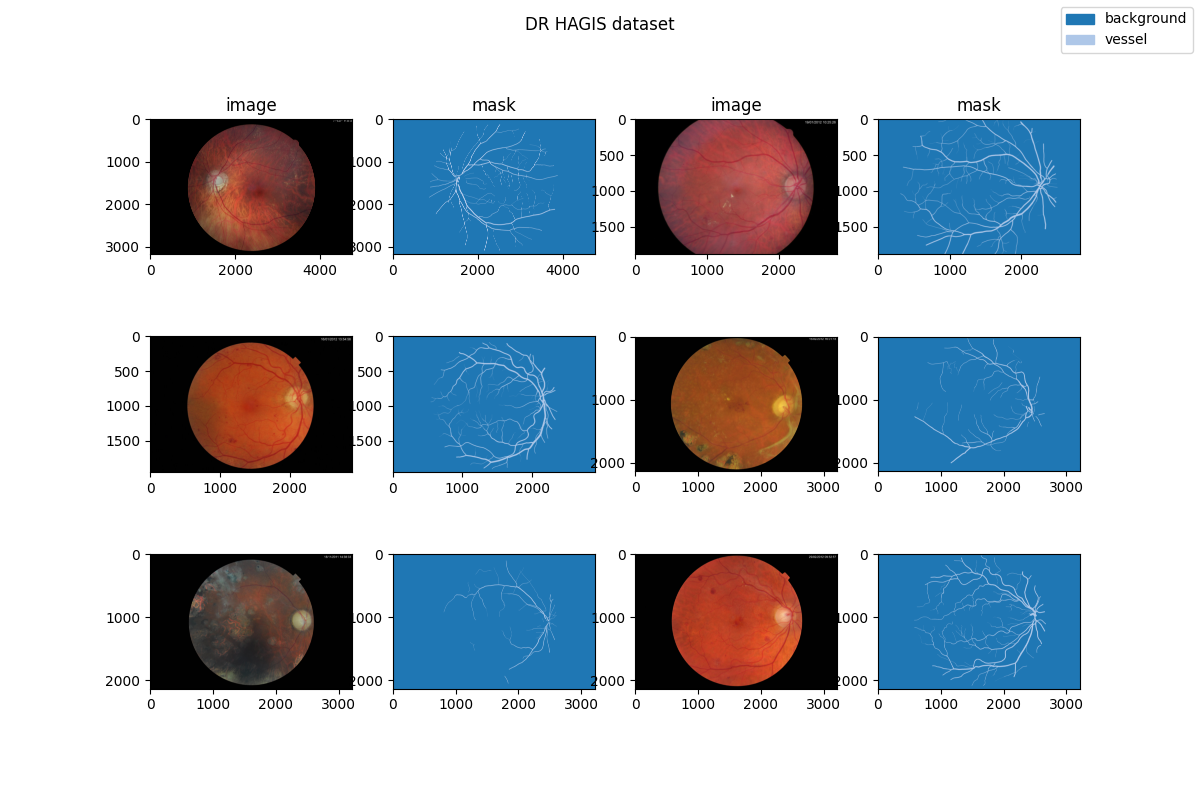 |
| 31 | + |
| 32 | +## Usage |
| 33 | + |
| 34 | +### Prerequisites |
| 35 | + |
| 36 | +- Python v3.8 |
| 37 | +- PyTorch v1.10.0 |
| 38 | +- [MIM](https://github.com/open-mmlab/mim) v0.3.4 |
| 39 | +- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 |
| 40 | +- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher |
| 41 | +- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5 |
| 42 | + |
| 43 | +All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `dr_hagis/` root directory, run the following line to add the current directory to `PYTHONPATH`: |
| 44 | + |
| 45 | +```shell |
| 46 | +export PYTHONPATH=`pwd`:$PYTHONPATH |
| 47 | +``` |
| 48 | + |
| 49 | +### Dataset preparing |
| 50 | + |
| 51 | +- download dataset from [here](https://paperswithcode.com/dataset/dr-hagis) and decompress data to path `'data/'`. |
| 52 | +- run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below. |
| 53 | +- run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set can't be obtained, we generate `train.txt` and `val.txt` from the training set randomly. |
| 54 | + |
| 55 | +```none |
| 56 | + mmsegmentation |
| 57 | + ├── mmseg |
| 58 | + ├── projects |
| 59 | + │ ├── medical |
| 60 | + │ │ ├── 2d_image |
| 61 | + │ │ │ ├── fundus_photography |
| 62 | + │ │ │ │ ├── dr_hagis |
| 63 | + │ │ │ │ │ ├── configs |
| 64 | + │ │ │ │ │ ├── datasets |
| 65 | + │ │ │ │ │ ├── tools |
| 66 | + │ │ │ │ │ ├── data |
| 67 | + │ │ │ │ │ │ ├── train.txt |
| 68 | + │ │ │ │ │ │ ├── val.txt |
| 69 | + │ │ │ │ │ │ ├── images |
| 70 | + │ │ │ │ │ │ │ ├── train |
| 71 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 72 | + │ │ │ │ | │ │ │ ├── ... |
| 73 | + │ │ │ │ | │ │ │ └── xxx.png |
| 74 | + │ │ │ │ │ │ ├── masks |
| 75 | + │ │ │ │ │ │ │ ├── train |
| 76 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 77 | + │ │ │ │ | │ │ │ ├── ... |
| 78 | + │ │ │ │ | │ │ │ └── xxx.png |
| 79 | +``` |
| 80 | + |
| 81 | +### Divided Dataset Information |
| 82 | + |
| 83 | +***Note: The table information below is divided by ourselves.*** |
| 84 | + |
| 85 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 86 | +| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 87 | +| background | 32 | 96.21 | 8 | 97.12 | - | - | |
| 88 | +| vessel | 32 | 3.79 | 8 | 2.88 | - | - | |
| 89 | + |
| 90 | +### Training commands |
| 91 | + |
| 92 | +Train models on a single server with one GPU. |
| 93 | + |
| 94 | +```shell |
| 95 | +mim train mmseg ./configs/${CONFIG_FILE} |
| 96 | +``` |
| 97 | + |
| 98 | +### Testing commands |
| 99 | + |
| 100 | +Test models on a single server with one GPU. |
| 101 | + |
| 102 | +```shell |
| 103 | +mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH} |
| 104 | +``` |
| 105 | + |
| 106 | +<!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models) |
| 107 | +
|
| 108 | +You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. --> |
| 109 | + |
| 110 | +## Dataset Citation |
| 111 | + |
| 112 | +If this work is helpful for your research, please consider citing the below paper. |
| 113 | + |
| 114 | +``` |
| 115 | +@article{holm2017dr, |
| 116 | + title={DR HAGIS—a fundus image database for the automatic extraction of retinal surface vessels from diabetic patients}, |
| 117 | + author={Holm, Sven and Russell, Greg and Nourrit, Vincent and McLoughlin, Niall}, |
| 118 | + journal={Journal of Medical Imaging}, |
| 119 | + volume={4}, |
| 120 | + number={1}, |
| 121 | + pages={014503--014503}, |
| 122 | + year={2017}, |
| 123 | + publisher={Society of Photo-Optical Instrumentation Engineers} |
| 124 | +} |
| 125 | +``` |
| 126 | + |
| 127 | +## Checklist |
| 128 | + |
| 129 | +- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. |
| 130 | + |
| 131 | + - [x] Finish the code |
| 132 | + |
| 133 | + - [x] Basic docstrings & proper citation |
| 134 | + |
| 135 | + - [ ] Test-time correctness |
| 136 | + |
| 137 | + - [x] A full README |
| 138 | + |
| 139 | +- [ ] Milestone 2: Indicates a successful model implementation. |
| 140 | + |
| 141 | + - [ ] Training-time correctness |
| 142 | + |
| 143 | +- [ ] Milestone 3: Good to be a part of our core package! |
| 144 | + |
| 145 | + - [ ] Type hints and docstrings |
| 146 | + |
| 147 | + - [ ] Unit tests |
| 148 | + |
| 149 | + - [ ] Code polishing |
| 150 | + |
| 151 | + - [ ] Metafile.yml |
| 152 | + |
| 153 | +- [ ] Move your modules into the core package following the codebase's file hierarchy structure. |
| 154 | + |
| 155 | +- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
0 commit comments