|
| 1 | +# CoNIC: Colon Nuclei Identification and Counting Challenge |
| 2 | + |
| 3 | +## Description |
| 4 | + |
| 5 | +This project supports **`CoNIC: Colon Nuclei Identification and Counting Challenge`**, which can be downloaded from [here](https://drive.google.com/drive/folders/1il9jG7uA4-ebQ_lNmXbbF2eOK9uNwheb). |
| 6 | + |
| 7 | +### Dataset Overview |
| 8 | + |
| 9 | +Nuclear segmentation, classification and quantification within Haematoxylin & Eosin stained histology images enables the extraction of interpretable cell-based features that can be used in downstream explainable models in computational pathology (CPath). To help drive forward research and innovation for automatic nuclei recognition in CPath, we organise the Colon Nuclei Identification and Counting (CoNIC) Challenge. The challenge requires researchers to develop algorithms that perform segmentation, classification and counting of 6 different types of nuclei within the current largest known publicly available nuclei-level dataset in CPath, containing around half a million labelled nuclei. |
| 10 | + |
| 11 | +### Task Information |
| 12 | + |
| 13 | +The CONIC challenge has 2 tasks: |
| 14 | + |
| 15 | +- Task 1: Nuclear segmentation and classification. |
| 16 | + |
| 17 | +The first task requires participants to segment nuclei within the tissue, while also classifying each nucleus into one of the following categories: epithelial, lymphocyte, plasma, eosinophil, neutrophil or connective tissue. |
| 18 | + |
| 19 | +- Task 2: Prediction of cellular composition. |
| 20 | + |
| 21 | +For the second task, we ask participants to predict how many nuclei of each class are present in each input image. |
| 22 | + |
| 23 | +The output of Task 1 can be directly used to perform Task 2, but these can be treated as independent tasks. Therefore, if it is preferred, prediction of cellular composition can be treated as a stand alone regression task. |
| 24 | + |
| 25 | +***NOTE:We only consider `Task 1` in the following sections.*** |
| 26 | + |
| 27 | +### Original Statistic Information |
| 28 | + |
| 29 | +| Dataset name | Anatomical region | Task type | Modality | Num. Classes | Train/Val/Test Images | Train/Val/Test Labeled | Release Date | License | |
| 30 | +| -------------------------------------------------------- | ----------------- | ------------ | -------------- | ------------ | --------------------- | ---------------------- | ------------ | ------------------------------------------------------------------------------------------------------------ | |
| 31 | +| [CoNIC202](https://conic-challenge.grand-challenge.org/) | abdomen | segmentation | histopathology | 7 | 4981/-/- | yes/-/- | 2022 | [Attribution-NonCommercial-ShareAlike 4.0 International](https://creativecommons.org/licenses/by-nc-sa/4.0/) | |
| 32 | + |
| 33 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 34 | +| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 35 | +| background | 4981 | 83.97 | - | - | - | - | |
| 36 | +| neutrophil | 1218 | 0.13 | - | - | - | - | |
| 37 | +| epithelial | 4256 | 10.31 | - | - | - | - | |
| 38 | +| lymphocyte | 4473 | 1.85 | - | - | - | - | |
| 39 | +| plasma | 3316 | 0.55 | - | - | - | - | |
| 40 | +| eosinophil | 1456 | 0.1 | - | - | - | - | |
| 41 | +| connective | 4613 | 3.08 | - | - | - | - | |
| 42 | + |
| 43 | +Note: |
| 44 | + |
| 45 | +- `Pct` means percentage of pixels in this category in all pixels. |
| 46 | + |
| 47 | +### Visualization |
| 48 | + |
| 49 | +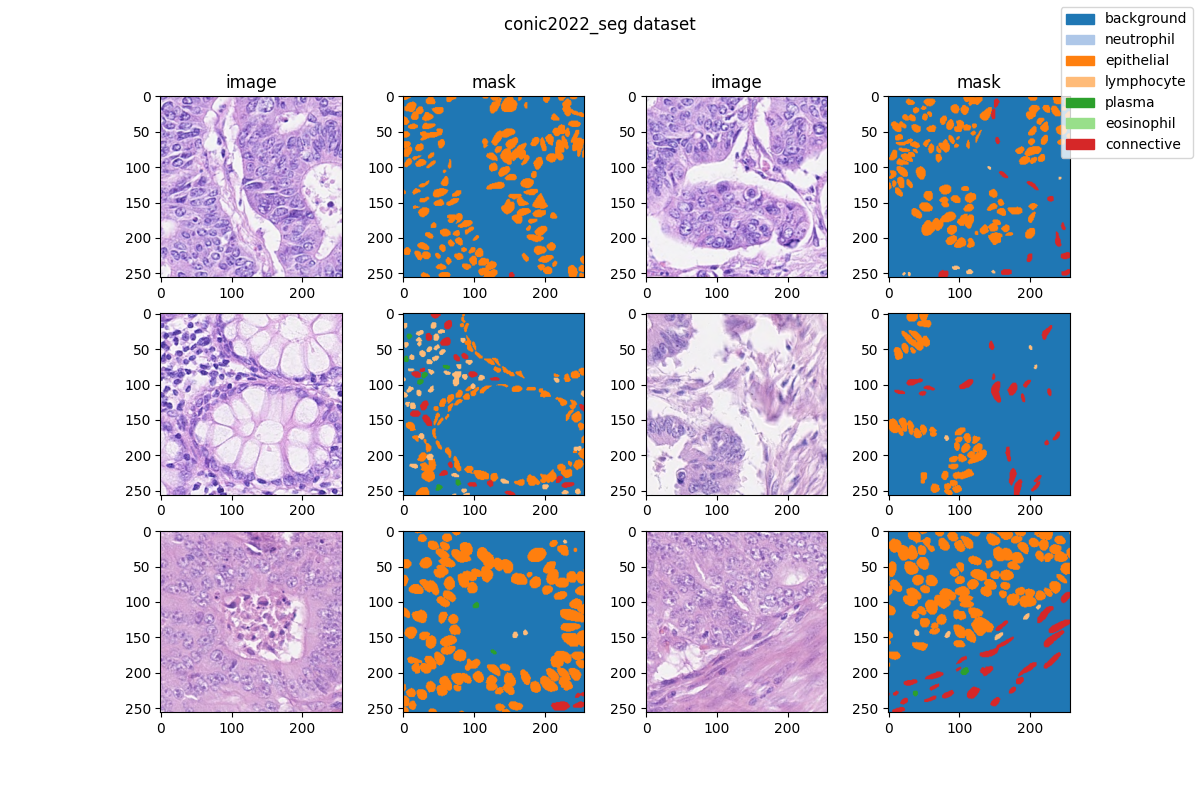 |
| 50 | + |
| 51 | +### Prerequisites |
| 52 | + |
| 53 | +- Python v3.8 |
| 54 | +- PyTorch v1.10.0 |
| 55 | +- pillow(PIL) v9.3.0 |
| 56 | +- scikit-learn(sklearn) v1.2.0 |
| 57 | +- [MIM](https://github.com/open-mmlab/mim) v0.3.4 |
| 58 | +- [MMCV](https://github.com/open-mmlab/mmcv) v2.0.0rc4 |
| 59 | +- [MMEngine](https://github.com/open-mmlab/mmengine) v0.2.0 or higher |
| 60 | +- [MMSegmentation](https://github.com/open-mmlab/mmsegmentation) v1.0.0rc5 |
| 61 | + |
| 62 | +All the commands below rely on the correct configuration of `PYTHONPATH`, which should point to the project's directory so that Python can locate the module files. In `conic2022_seg/` root directory, run the following line to add the current directory to `PYTHONPATH`: |
| 63 | + |
| 64 | +```shell |
| 65 | +export PYTHONPATH=`pwd`:$PYTHONPATH |
| 66 | +``` |
| 67 | + |
| 68 | +### Dataset preparing |
| 69 | + |
| 70 | +- download dataset from [here](https://drive.google.com/drive/folders/1il9jG7uA4-ebQ_lNmXbbF2eOK9uNwheb/) and move data to path `'data/CoNIC_Challenge'`. The directory should be like: |
| 71 | + ```shell |
| 72 | + data/CoNIC_Challenge |
| 73 | + ├── README.txt |
| 74 | + ├── by-nc-sa.md |
| 75 | + ├── counts.csv |
| 76 | + ├── images.npy |
| 77 | + ├── labels.npy |
| 78 | + └── patch_info.csv |
| 79 | + ``` |
| 80 | +- run script `"python tools/prepare_dataset.py"` to format data and change folder structure as below. |
| 81 | +- run script `"python ../../tools/split_seg_dataset.py"` to split dataset and generate `train.txt`, `val.txt` and `test.txt`. If the label of official validation set and test set can't be obtained, we generate `train.txt` and `val.txt` from the training set randomly. |
| 82 | + |
| 83 | +```none |
| 84 | + mmsegmentation |
| 85 | + ├── mmseg |
| 86 | + ├── projects |
| 87 | + │ ├── medical |
| 88 | + │ │ ├── 2d_image |
| 89 | + │ │ │ ├── histopathology |
| 90 | + │ │ │ │ ├── conic2022_seg |
| 91 | + │ │ │ │ │ ├── configs |
| 92 | + │ │ │ │ │ ├── datasets |
| 93 | + │ │ │ │ │ ├── tools |
| 94 | + │ │ │ │ │ ├── data |
| 95 | + │ │ │ │ │ │ ├── train.txt |
| 96 | + │ │ │ │ │ │ ├── val.txt |
| 97 | + │ │ │ │ │ │ ├── images |
| 98 | + │ │ │ │ │ │ │ ├── train |
| 99 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 100 | + │ │ │ │ | │ │ │ ├── ... |
| 101 | + │ │ │ │ | │ │ │ └── xxx.png |
| 102 | + │ │ │ │ │ │ ├── masks |
| 103 | + │ │ │ │ │ │ │ ├── train |
| 104 | + │ │ │ │ | │ │ │ ├── xxx.png |
| 105 | + │ │ │ │ | │ │ │ ├── ... |
| 106 | + │ │ │ │ | │ │ │ └── xxx.png |
| 107 | +``` |
| 108 | + |
| 109 | +### Divided Dataset Information |
| 110 | + |
| 111 | +***Note: The table information below is divided by ourselves.*** |
| 112 | + |
| 113 | +| Class Name | Num. Train | Pct. Train | Num. Val | Pct. Val | Num. Test | Pct. Test | |
| 114 | +| :--------: | :--------: | :--------: | :------: | :------: | :-------: | :-------: | |
| 115 | +| background | 3984 | 84.06 | 997 | 83.65 | - | - | |
| 116 | +| neutrophil | 956 | 0.12 | 262 | 0.13 | - | - | |
| 117 | +| epithelial | 3400 | 10.26 | 856 | 10.52 | - | - | |
| 118 | +| lymphocyte | 3567 | 1.83 | 906 | 1.96 | - | - | |
| 119 | +| plasma | 2645 | 0.55 | 671 | 0.56 | - | - | |
| 120 | +| eosinophil | 1154 | 0.1 | 302 | 0.1 | - | - | |
| 121 | +| connective | 3680 | 3.08 | 933 | 3.08 | - | - | |
| 122 | + |
| 123 | +### Training commands |
| 124 | + |
| 125 | +Train models on a single server with one GPU. |
| 126 | + |
| 127 | +```shell |
| 128 | +mim train mmseg ./configs/${CONFIG_FILE} |
| 129 | +``` |
| 130 | + |
| 131 | +### Testing commands |
| 132 | + |
| 133 | +Test models on a single server with one GPU. |
| 134 | + |
| 135 | +```shell |
| 136 | +mim test mmseg ./configs/${CONFIG_FILE} --checkpoint ${CHECKPOINT_PATH} |
| 137 | +``` |
| 138 | + |
| 139 | +<!-- List the results as usually done in other model's README. [Example](https://github.com/open-mmlab/mmsegmentation/tree/dev-1.x/configs/fcn#results-and-models) |
| 140 | +
|
| 141 | +You should claim whether this is based on the pre-trained weights, which are converted from the official release; or it's a reproduced result obtained from retraining the model in this project. --> |
| 142 | + |
| 143 | +## Organizers |
| 144 | + |
| 145 | +- Simon Graham (TIA, PathLAKE) |
| 146 | +- Mostafa Jahanifar (TIA, PathLAKE) |
| 147 | +- Dang Vu (TIA) |
| 148 | +- Giorgos Hadjigeorghiou (TIA, PathLAKE) |
| 149 | +- Thomas Leech (TIA, PathLAKE) |
| 150 | +- David Snead (UHCW, PathLAKE) |
| 151 | +- Shan Raza (TIA, PathLAKE) |
| 152 | +- Fayyaz Minhas (TIA, PathLAKE) |
| 153 | +- Nasir Rajpoot (TIA, PathLAKE) |
| 154 | + |
| 155 | +TIA: Tissue Image Analytics Centre, Department of Computer Science, University of Warwick, United Kingdom |
| 156 | + |
| 157 | +UHCW: Department of Pathology, University Hospitals Coventry and Warwickshire, United Kingdom |
| 158 | + |
| 159 | +PathLAKE: Pathology Image Data Lake for Analytics Knowledge & Education, , University Hospitals Coventry and Warwickshire, United Kingdom |
| 160 | + |
| 161 | +## Dataset Citation |
| 162 | + |
| 163 | +If this work is helpful for your research, please consider citing the below paper. |
| 164 | + |
| 165 | +``` |
| 166 | +@inproceedings{graham2021lizard, |
| 167 | + title={Lizard: A large-scale dataset for colonic nuclear instance segmentation and classification}, |
| 168 | + author={Graham, Simon and Jahanifar, Mostafa and Azam, Ayesha and Nimir, Mohammed and Tsang, Yee-Wah and Dodd, Katherine and Hero, Emily and Sahota, Harvir and Tank, Atisha and Benes, Ksenija and others}, |
| 169 | + booktitle={Proceedings of the IEEE/CVF International Conference on Computer Vision}, |
| 170 | + pages={684--693}, |
| 171 | + year={2021} |
| 172 | +} |
| 173 | +@article{graham2021conic, |
| 174 | + title={Conic: Colon nuclei identification and counting challenge 2022}, |
| 175 | + author={Graham, Simon and Jahanifar, Mostafa and Vu, Quoc Dang and Hadjigeorghiou, Giorgos and Leech, Thomas and Snead, David and Raza, Shan E Ahmed and Minhas, Fayyaz and Rajpoot, Nasir}, |
| 176 | + journal={arXiv preprint arXiv:2111.14485}, |
| 177 | + year={2021} |
| 178 | +} |
| 179 | +``` |
| 180 | + |
| 181 | +## Checklist |
| 182 | + |
| 183 | +- [x] Milestone 1: PR-ready, and acceptable to be one of the `projects/`. |
| 184 | + |
| 185 | + - [x] Finish the code |
| 186 | + |
| 187 | + - [x] Basic docstrings & proper citation |
| 188 | + |
| 189 | + - [x] A full README |
| 190 | + |
| 191 | +- [ ] Milestone 2: Indicates a successful model implementation. |
| 192 | + |
| 193 | + - [ ] Training-time correctness |
| 194 | + |
| 195 | +- [ ] Milestone 3: Good to be a part of our core package! |
| 196 | + |
| 197 | + - [ ] Type hints and docstrings |
| 198 | + |
| 199 | + - [ ] Unit tests |
| 200 | + |
| 201 | + - [ ] Code polishing |
| 202 | + |
| 203 | + - [ ] Metafile.yml |
| 204 | + |
| 205 | +- [ ] Move your modules into the core package following the codebase's file hierarchy structure. |
| 206 | + |
| 207 | +- [ ] Refactor your modules into the core package following the codebase's file hierarchy structure. |
0 commit comments